Native American Branch
Q-M3 > Q-M848 are the main native american lineages. TMRCA of Q-M3 is 12600 YBP. However a parallel but upstream lineage Q-M1107 TMRCA 15400 YBP is also found in natives. Both Q-M3 and Q-M1107 come from Q-L54 ultimately. Q-M3 comes from Q-M930 but the latter has some European exclusive lineages too. Q-M3 is also known as Q1a2a1a1.
Indian Branches
1.
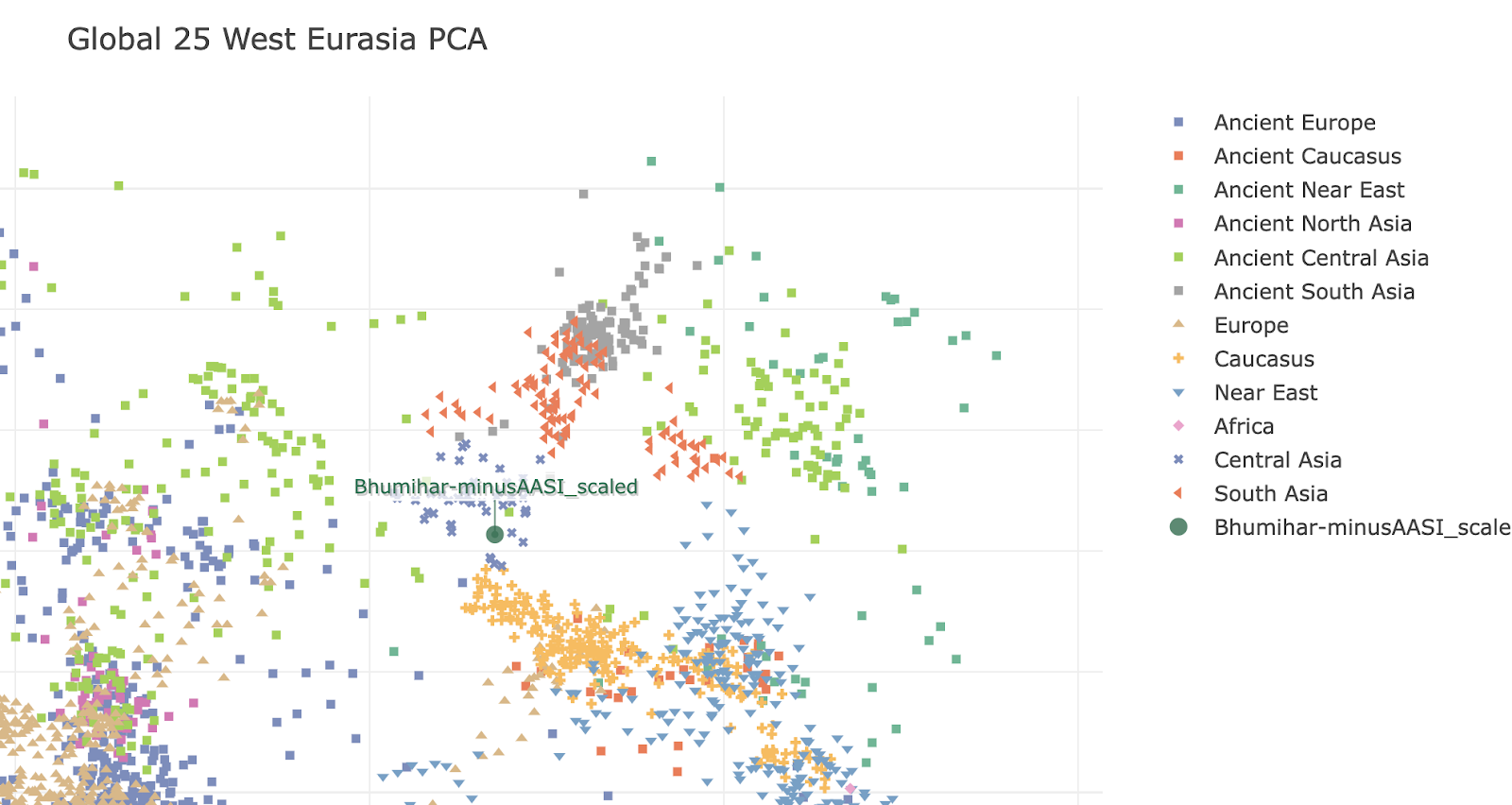
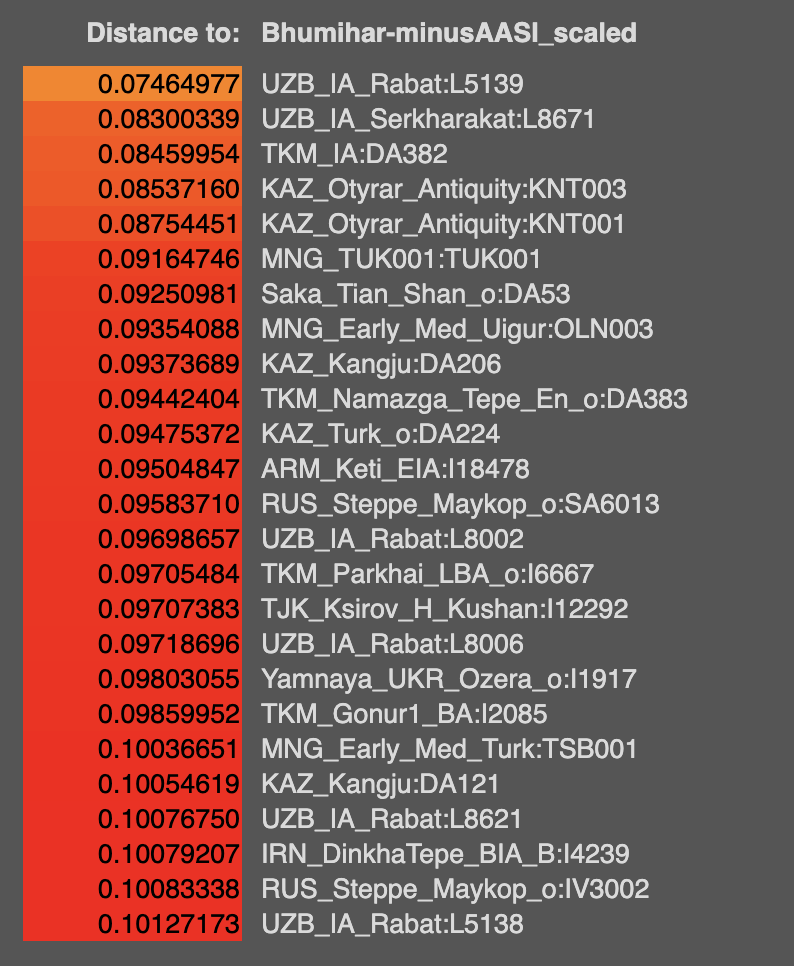
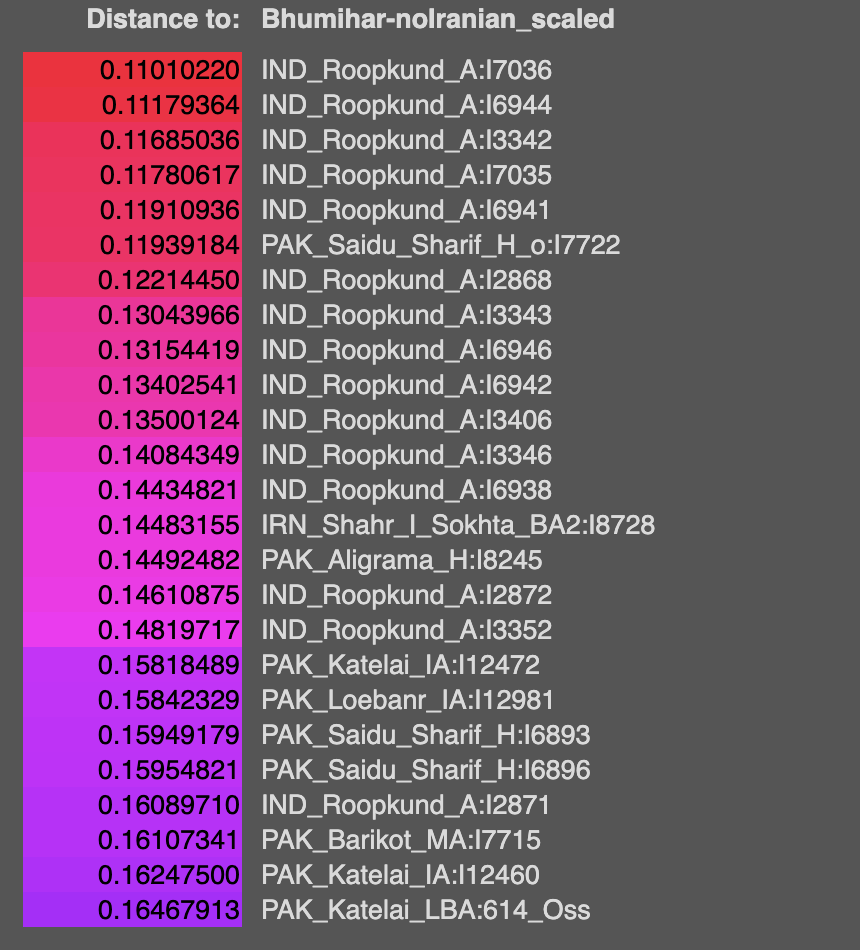
Q-L275 or Q3 is found in India. Oldest sample with it is an Afanasievo Altai one C2034 ~ 2500 BCE from Kumar et al 2022. Subclade is Q-NGQ1. Then a few ones from Xinjiang, one Chemurchek and the Sappali Tepe outlier that is Indian like in admixture I7493 context dated 2000-1600 BCE is also Q3 (Q-Z19128 / TMRCA 3699 BCE). Then we get 2 people from Loebanr who carry it (I13228, I5400) dated to approx 1000-800 BCE. They are on the a clade downstream from Q-Z19128. Abusanteer 7 from Kumar et al or C4272 also carries Q-Y2265 (the sample is dated to 700-200 BCE). One of the historical Saidu Sharif samples I7718 also carries Q-L275, subclade is Q-Z19128 same as the Sappali Tepe outlier. Finally, Roopkund 53 or I6939 also carries Q-L275 > Q-L245.
Now, looking at the modern data for Q-L275 at
Y-full and
FT-DNA. Q-Z19128 is only shared between Turkics and South Asians. Probably a non-WSH/Afanasievo derived line? Otoh, Q-NGQ1 > Q-M378 is found in Europeans as well as South-Central Asians. It's also found in one Afanasievo sample, and one Xinjiang EBA steppe sample. The Abusanteer 7 clade Q-Y2265 is found in a shit load of South Asians and one Turk, one Chinese Tajik on Y-full.
On Y-full, Q-Y2200 2500ybp formed, TMRCA 1500 ybp seems to be a uniquely Balto-Slavic clade, found in some Israeli Yiddish speakers too. This is the only European exclusive branch of Q-L275 I've found there.
2. The main East Asian Q-clade is
Q-Y647 (Q > Q-L472 > Q-Y570 > Q-M120 > Q-Y647) and it has a TMRCA of 4000 BCE.
relevant papers