In South Asian genetics, we suffer from a paucity of ancient DNA to study. We have about 100 samples from Pakistan, Gandhara to be precise. These samples range from the Late-Bronze Age to the Medieval Historic period. Then, we have about two dozen samples from Roopkund Lake that are generally from antiquity but going up to the middle ages. This is all we have to work with. So let's analyze the profile of the Swat Protohistoric samples first.
One thing I would like to discuss before we begin is the presence of BMAC/Oxus/Turanian Bronze Age DNA in these Swat samples. Many people take G25 models too seriously and believe there is upwards of 30-45% Oxus ancestry in the Gandhara samples. I think this is entirely wrong, as formal tools such as qpAdm reject BMAC decisively or give very little amounts of it (2-5%)
To illustrate my point, take a look at a G25 profile of the samples with BMAC + Sintashta.
30% BMAC in Loebanr_IA and 25% BMAC in Katelai_IA! Keep this ridiculously high number in mind. Now, let us see what qpAdm says.
weights
# A tibble: 3 × 5
target left weight se z
<chr> <chr> <dbl> <dbl> <dbl>
1 Pakistan_Katelai_IA Russia_MLBA_Sintashta 0.163 0.0197 8.29
2 Pakistan_Katelai_IA Indus_Gonur 0.816 0.0395 20.7
3 Pakistan_Katelai_IA Uzbekistan_Dzharkutan_BA_1 0.0211 0.0459 0.459
2.1% BMAC/Oxus! down from the ridiculous 25% given before.
Now, let's look at Loebanr_IA
$weights
# A tibble: 3 × 5
target left weight se z
<chr> <chr> <dbl> <dbl> <dbl>
1 Pakistan_Loebanr_IA Russia_MLBA_Sintashta 0.176 0.0194 9.03
2 Pakistan_Loebanr_IA Indus_Gonur 0.832 0.0404 20.6
3 Pakistan_Loebanr_IA Uzbekistan_Dzharkutan_BA_1 -0.00746 0.0463 -0.161
Pure BMAC is literally rejected as a source. Loebanr_IA gets modeled purely as Indus_Gonur + Sintashta. This is doubt enough to make us deeply suspicious of G25 runs for South Asians that use pure BMAC as a source. What's likely happening here is an overfit + affinity to BMAC based on higher proportions of ANF/CHG ancestry that are missing from the Indus Valley samples we currently have. Increased sampling of the IVC might fix this. The only way Indians get any BMAC is indirectly, via Steppe sources mixed with BMAC like Dashti Kozy or later historic ones like Kangju or the Iron Age Yaz sample TKM_IA (Takhirbai_IA). This indirect BMAC ancestry peaks in North-Western populations at perhaps 15%, nothing more. G25 however is not able to differentiate IVC from BMAC at the moment, so all calculators using BMAC are not very useful at the moment.
This is indeed the conclusion that Narasimhan et al 2019 came to also.
Therefore, here is a model of the Swat Protohistoric samples without BMAC/Oxus as a source.
Kumsay here serves as proxy of the kind of Central Asian ancestry the Andronovo Pastoralists might have picked up on there way to the subcontinent. Using the same model, here is what we get for modern North-West Indian and some Pakistani populations.
Modern samples have more Steppe than most of the ancient ones, but the devil lies in the detail and in population structure. The modern samples are fairly homogeneous (barring the Punjabi_Lahore set which is a mix of all kinds of Pakistani castes). Are the ancient sample sets also homogeneous? Let's see.
Indeed, what we find is that the Iron Age samples are not homogeneous, which makes sense. The modern samples are from stratified and endogamous Indian caste groups that have avoided intermarrying each other for millenia, the ancient ones are a graveyard dump of all sorts of people. We see that some of the Loebanr_IA samples are as low as 5% Steppe (I12981, I12134) while some of the Katelai_IA are as low as 2% Steppe (I12446, I12470, I12460) while some are as high as 28% Steppe (I12141). The Udegram_IA, Saidu_Sharif_H and Butkara_IA ones are far more homogeneous. Saidu Sharif_H has one outlier that is like 33-35% Steppe and one outlier that is like <5% Steppe.
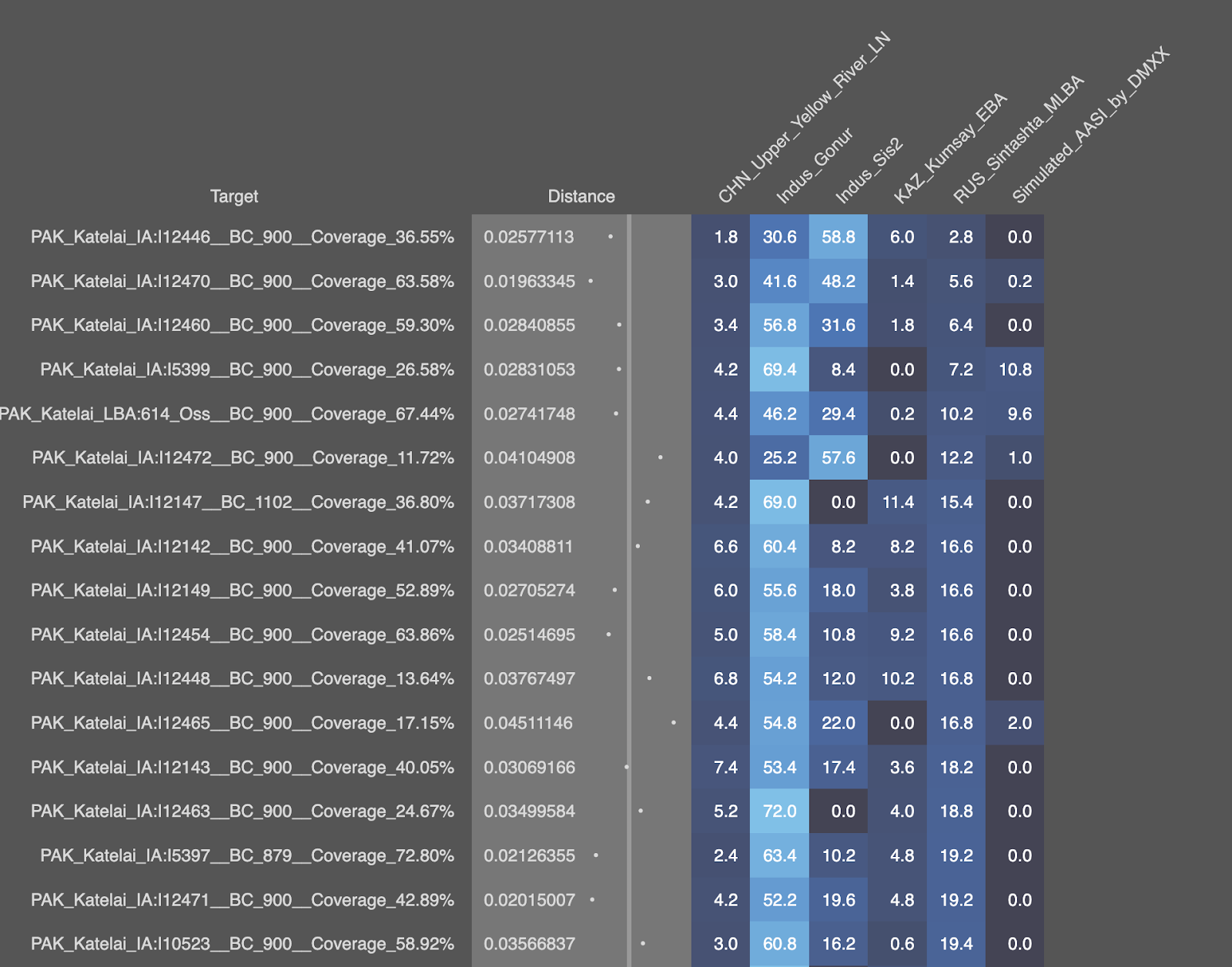
Here are all the runs posted.
For Katelai_IA, will split it in 2 screenshots.
Similarly for Loebanr_IA due to large number of samples.
Even after accounting for most of the outliers, we can see that in general, the Iron/Bronze Age and Historic samples are about 5-8% lower Steppe_MLBA than modern North Westerners. Perhaps this points to different waves of migration and a different wave of Indo-Aryans giving rise to modern North Westerners (with differences amongst themselves too). This can be confirmed by Y-DNA to a regard, where tribes such as Khatris get 65% R1a-Z93 with most of it being the Indian L-657 while the Swat Protohistoric samples are maxxed out in J2, E1b, L-M20 with barely any R-Z93s. Hence, clearly different paternal lineages gave rise to lot of these tribes. Those lineages must've different in their exact autosomal profile too, even if it was very similar. For more on the Swat haplogroups, see this post of mine